R is commonly known as a programming language used by statisticians and data scientists. While this was largely accurate in the past, the adaptability offered by R’s packages has transformed it into a more versatile language. Open sourced in 1995, R has experienced repositories of R packages are constantly growing since then. Nevertheless, R remains heavily data-centric compared to languages such as Python.
Among data types, tabular data stands out due to its widespread use. This format, resembling database tables, allows each column to hold a different data type. The efficiency of processing this data type is critical for many applications.

This article aims to demonstrate how to efficiently transform tabular data in R. Many R users, even those familiar with machine learning, are unaware that data munging can be achieved faster in R without relying on external tools.
High-Performance Tabular Data Transformation with R
Introduced in 1997, the data.frame class in base R, inherited from S-PLUS, stores data in memory in a column-oriented structure, unlike many databases that utilize row-by-row storage. This approach optimizes cache efficiency for column-based operations common in analytics. Additionally, R, despite being a functional programming language, doesn’t enforce this paradigm on developers. These advantages are effectively leveraged by the data.table R package, available in the CRAN repository. This package excels in grouping operations and memory efficiency by carefully materializing intermediate data subsets, generating only the columns required for a specific task. It also eliminates redundant copies through its reference semantics when adding or updating columns. First released in April 2006, the package significantly enhanced data.frame performance. The initial package description stated:
This package is intentionally minimal. Its sole purpose is to address the “white book” specification that
data.framemust have row names. It introduces thedata.tableclass, functionally similar todata.framebut with significantly reduced memory usage (up to 10 times less) and faster creation and copying (up to 10 times faster). Additionally, it enablessubset()andwith()-like expressions within[]. Most of the code is adapted from base functions, removing row name manipulation.
Both data.frame and data.table have undergone improvements since then, but data.table continues to outperform base R significantly. In fact, it’s considered one of the fastest open-source data manipulation tools available, rivaling tools like Python Pandas and columnar databases or big data applications like Spark. Although benchmarks for its performance in distributed shared infrastructure are pending, its capacity to handle up to two billion rows on a single instance is promising. Its exceptional performance is attributed to its functionalities. Furthermore, ongoing efforts to parallelize time-consuming operations for incremental performance gains suggest a clear path for future optimization.
Illustrative Examples of Data Transformation
The interactive nature of R simplifies learning by allowing us to examine results at each step. Before proceeding, let’s install the data.table package from CRAN:
| |
Helpful Tip: Access the manual for any function by prefixing its name with a question mark, for instance, ?install.packages.
Importing Data into R
R offers numerous packages for extracting data from various formats and databases, often including native drivers. In this example, we’ll load data from a CSV file, a prevalent format for raw tabular data. The file used in these examples is available here. We can rely on the highly optimized fread function for efficient CSV reading.
To utilize any function from a package, we need to load it using the library call:
| |
| |
If our data requires reshaping from long-to-wide or wide-to-long formats (also known as pivot and unpivot) for further processing, we can refer to the ?dcast and ?melt functions from the reshape2 package. However, data.table provides faster and more memory-efficient alternatives for data.table/data.frame objects.
Querying Data Using the data.table Syntax
If You Are Accustomed to data.frame
Querying data.table closely resembles querying data.frame. When filtering in the i argument, we can directly use column names without the need for the $ sign, as in df[df$col > 1, ]. The j argument accepts an expression to be evaluated within the scope of our data.table. To pass a non-expression j argument, use with=FALSE. The third argument, absent in the data.frame method, defines the groups, causing the expression in j to be evaluated by groups.
| |
If You Are Familiar with Databases
Querying data.table shares similarities with SQL queries. DT in the example below symbolizes a data.table object and corresponds to SQL’s FROM clause.
| |
Sorting Rows and Reordering Columns
Sorting data is essential for time series analysis and crucial for data extraction and presentation. Sorting can be achieved by providing the integer vector of row order to the i argument, similar to data.frame. The first argument in the query order(carrier, -dep_delay) sorts data in ascending order based on the carrier field and descending order based on the dep_delay measure. The second argument j, as explained earlier, specifies the columns (or expressions) to be returned and their order.
| |
| |
To reorder data by reference instead of querying data in a specific order, we use the set* functions.
| |
| |
In most cases, we only need either the original dataset or the ordered/sorted dataset. R, like other functional programming languages, by default returns sorted data as a new object, potentially doubling memory usage compared to sorting by reference.
Subset Queries
Let’s create a subset dataset for flights originating from “JFK” and departing between June and September. In the second argument, we limit the results to the listed columns, including a calculated variable sum_delay.
| |
| |
When subsetting a dataset based on a single column, data.table automatically creates an index for that column. This results in real-time responses for subsequent filtering operations on that column.
Updating Datasets
Adding a new column by reference is performed using the := operator, which assigns a variable to the dataset in place. This avoids copying the dataset in memory, eliminating the need to assign results to a new variable.
| |
| |
To add multiple variables simultaneously, use the syntax DT[, := (sum_delay = arr_delay + dep_delay)], similar to .(sum_delay = arr_delay + dep_delay) when querying the dataset.
Sub-assignment by reference, updating specific rows in place, is achieved by combining the i argument with the assignment operator.
| |
| |
Aggregating Data
Data aggregation is performed by providing the third argument by to the square brackets. The j argument should contain aggregate function calls to perform the aggregation. The .N symbol used in the j argument represents the total number of observations within each group. As previously mentioned, aggregates can be combined with row subsets and column selections.
| |
| |
Comparing a row’s value to its group aggregate is a common requirement. In SQL, we use aggregates over partition by: AVG(arr_delay) OVER (PARTITION BY carrier, month).
| |
| |
To update the table with aggregates by reference instead of querying, use the := operator. This avoids copying the dataset in memory, eliminating the need to assign results to a new variable.
| |
| |
Joining Datasets
In base R, joining and merging datasets are treated as specialized subset operations. The first square bracket argument i specifies the dataset to join with. For each row in the dataset provided to i, matching rows are found in the dataset being queried using [. To retain only matching rows (inner join), pass an additional argument nomatch = 0L. The on argument designates the columns on which to join the datasets.
| |
| |
| |
| |
| |
| |
Note that due to consistency with base R subsetting, the default outer join is a RIGHT OUTER join. For a LEFT OUTER join, swap the table order, as demonstrated in the example above. The merge method of data.table offers fine-grained control over join behavior, utilizing the same API as the base R merge function for data.frame.
Efficiently look up columns from another dataset using the := operator in the j argument while joining. This adds a column by reference from the joined dataset, avoiding in-memory data copying.
| |
| |
For aggregating while joining, use by = .EACHI. This performs a join without materializing intermediate results, applying aggregates on the fly for memory efficiency.
Rolling joins, uncommon but well-suited for ordered data, are ideal for processing temporal data and time series. They roll matches in the join condition to the next matching value. Utilize rolling joins by providing the roll argument during the join operation.
Fast overlap join joins datasets based on periods, handling overlaps using operators like any, within, start, and end.
Currently, the non-equi join feature for joining datasets using non-equal conditions is being developed.
Profiling Data
When exploring a dataset, gathering technical information can provide insights into data quality.
Descriptive Statistics
| |
| |
Cardinality
Determine the uniqueness of data using the uniqueN function on each column. The object .SD in the following query represents the Subset of the Data.table:
| |
| |
NA Ratio
Calculate the ratio of missing values (NA in R, NULL in SQL) for each column by providing the desired function to apply to every column.
| |
| |
Exporting Data
The data.table package also provides functionality for Fast export tabular data to CSV format.
| |
| |
At the time of writing, the fwrite function is not yet available on CRAN. To use it, install data.table development version; otherwise, the base R write.csv function can be used, but it might be slow.
Further Resources
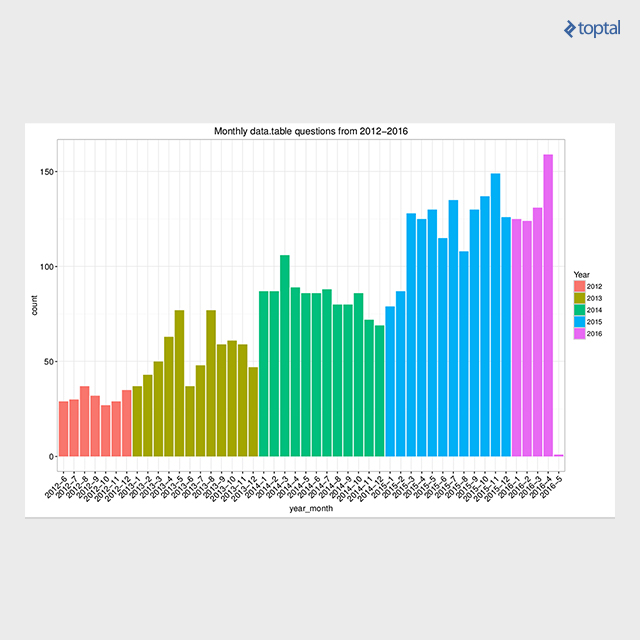
Ample resources are available, including function manuals and package vignettes offering subject-specific tutorials. These resources can be found on the Getting started page. The Presentations page lists over 30 materials (slides, videos, etc.) from data.table presentations worldwide. Additionally, a thriving community provides support, with over 4,000 questions tagged with data.table on Stack Overflow, boasting a high answer rate (91.9%). The plot below depicts the trend of data.table tagged questions on Stack Overflow over time.
Conclusion
This article presents select examples for efficient tabular data transformation in R using the data.table package. For concrete performance figures, refer to reproducible benchmarks. My blog post, Solve common R problems efficiently with data.table, summarizes data.table solutions for the top 50 rated Stack Overflow questions related to the R language, providing numerous figures and reproducible code. The data.table package employs native implementations of fast radix ordering for grouping and binary search for efficient subsets/joins. Radix ordering was incorporated into base R from version 3.3.0 onwards. Recently, this algorithm was implemented and parallelized within the H2O machine learning platform for use across H2O clusters, further enhancing its capabilities enabling efficient big joins on 10B x 10B rows.